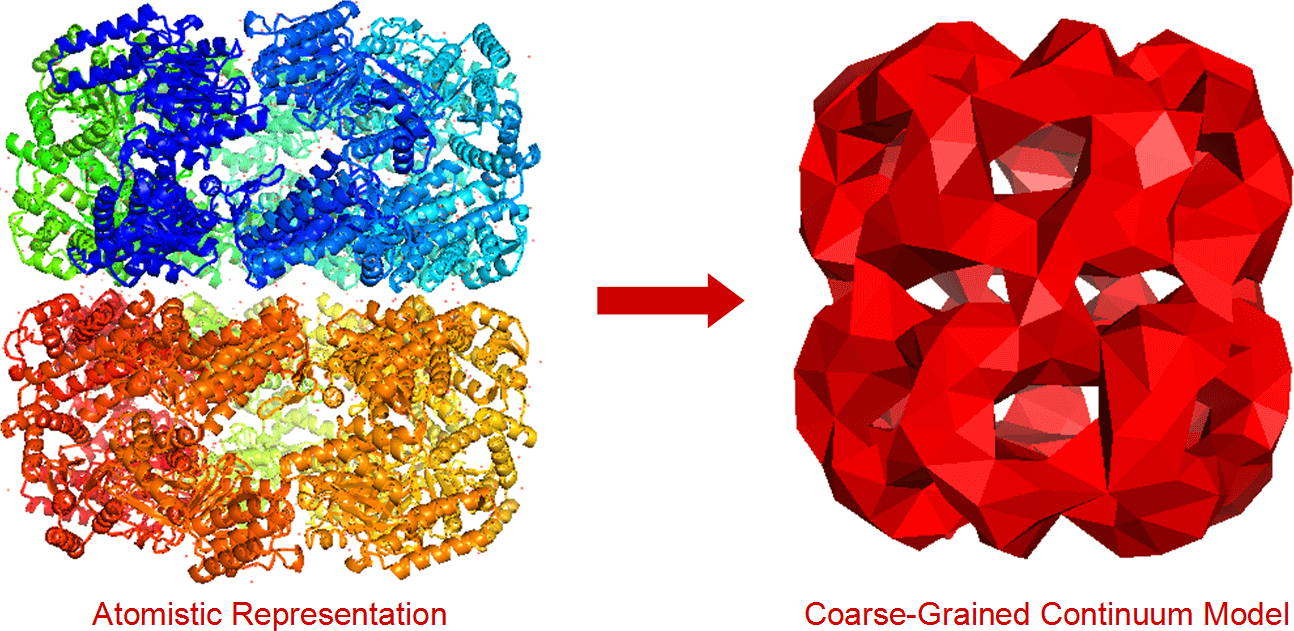
Fluctuating Finite Element Analysis is a new molecular modelling algorithm, designed to support systems that are larger and more complex than those modelled by atomistic molecular dynamics. Instead of modelling biological systems as a collection of connected atoms, it models them as 3D volumes comprised of tetrahedrons. Unlike previous coarse-grained models, the models FFEA generates are visco-elastic continuum solids. Unlike other applications of Finite Element Analysis, these systems are subject to thermal fluctuations.
This technique has the potential to model large, complex systems, made of many molecules, and complex processes at the frontiers of molecular biology. As it does not not require an atomistic level of detail, it can also be used to simulate biological molecules that cannot be imaged using X-ray crystallography.
Features
- Protein-protein interactions:
- A 6-12 Lennard-Jones Potential.
- A repulsive potential that is proportional to the overlapping volume.
- Specific interactions defined using [precomputed potentials](https://ffea.readthedocs.io/en/latest/fmApproach.html).
- Kinetic state changes can be simulated together with the continuum model to account for conformational changes and binding events.
- Conversion tools for EM density data and atomistic structures into FFEA simulations.
- A plugin for PyMOL, allowing the visualisation of FFEA systems and trajectories.
- Initialisation and analysis tools available on the command line and under a Python API.
- KOBRA model for slender biological objects such as coiled-coils
- Lees-Edwards boundary conditions.
- An extremely high degree of reproducity - check out our integration tests!
- Distance restraints with harmonic potentials, such as springs.
Getting Started
FFEA is free to download and use under the GPLv3 software license. We provide binary releases, and building from source is relatively painless.
- Download the most recent binary release of FFEA from BitBucket, or compile from source by cloning the repository. For the latest bleeding-edge features, switch to the development branch instead.
- If you're compiling from source, install FFEA according to the instructions found in the installation guide.
- Once FFEA is installed, consult the first-time user tutorial. For KOBRA rods, try the rods tutorial instead.
Publications
Methodology:
- Welch R. J., Harris S. A., Harlen O. G. & Read D. J. "KOBRA: A Fluctuating Elastic Rod Model for Slender Biological Macromolecules" (2020), Soft Matter 16, 7544-7555.
- Solernou A., Hanson B., Richardson R. A., Welch R., Read D. J., Harlen O. G. and Harris S. A. "Fluctuating Finite Element Analysis (FFEA): A continuum mechanics software tool for mesoscale simulation of biomolecules" (2018), PLOS Comput. Biol. 14(3):e1005897.
- Oliver R., Read D. J., Harlen O. G. and Harris S. A. "A Stochastic finite element model for the dynamics of globular macromolecules" (2013), J. Comp. Phys. 239, 147-165.
- Patargias G. N., Harris S. A. and Harding J. "A demonstration of the inhomogeneity of the local dielectric response of proteins by molecular dynamics simulations." (2010), J. Chem. Phys. 132(23):235103.
Applications:
- Richardson R. A., Hanson B. S., Read D. J., Harlen O. G. & Harris S. A. "Exploring the dynamics of flagellar dynein within the axoneme with Fluctuating Finite Element Analysis" (2020), Q. Rev. Biophys. 53, E9.
- Lee S. C., Collins R., Lin Y., Jamshad M., Broughton C., Harris S. A., Hanson B. S., Tognoloni C., Parslow R. A., Terry A. E., Rodger A., Smith C. J., Edler K. J., Ford R., Roper D. I. & Dafforn T. R. "Nano-encapsulated Escherichia coli Divisome Anchor ZipA, and in Complex with FtsZ" (2019), Sci. Rep. 9(1):18712.
- Richardson R., Papachristos K., Read D. J., Harlen O. G., Harrison M. A., Paci E., Muench S. P. and Harris S. A "Understanding the apparent stator-rotor connections in the rotary ATPase family using coarse-grained computer modelling" (2014), Proteins: Struct., Funct., Bioinf. 82, 3298-3311.
Reviews:
- Gravett, M. S. C., Cocking R. C., Curd A. P., Harlen O., Leng J., Muench S. P., Peckham M., Read D. J., Rogers J. F., Welch R. C. and Harris S. A. "Moving in the mesoscale: Understanding the mechanics of cytoskeletal molecular motors by combining mesoscale simulations with imaging" (2022), WIREs Comput Mol Sci. 12:e1570.
- Gray A., Harlen O. G., Harris S. A., Khalid S., Leung Y. M., Lonsdale R., Mulholland A. J., Pearson A. R., Read D. J. and Richardson R. A. "In pursuit of an accurate spatial and temporal model of biomolecules at the atomistic level: a perspective on computer simulation" (2015), Acta Crystallogr. D 71, 162-172.
- Oliver R., Richardson R. A., Hanson B., Kendrick K., Read D. J., Harlen O. G. and Harris S. A. "Modelling the Dynamic Architecture of Biomaterials Using Continuum Mechanics" (2014), Protein Modelling, Náray-Szabó G. (Ed.), Springer International Publishing, 175-197.
- Hanson B., Richardson R., Oliver R., Read D. J., Harlen O. and Harris S. "Modelling biomacromolecular assemblies with continuum mechanics" (2015), Biochem. Soc. Trans. 43, 186-192.
Prerequisites
Essential:
- C and C++ compilers (GCC >=6.3.0 or Intel >=15). There is some C++ code written using the C++11 standard, and so CMake will ensure that you have a recent enough compiler. FFEA will compile with GCC 6.3.0 or Intel 15, but we suggest using an up to date compiler to get the best performance.
- CMake (>=2.8.11). Required for building FFEA.
- Python (2.7.X). Used to run the FFEA tools modules, in addition to unit and integration tests to verify that FFEA was correctly built. The NumPy, SciPy and Matplotlib libraries are required. FFEA tools is currently incompatible with Python 3, but an upgrade is in progress.
Recommended:
- PyMOL(>=1.8, though 1.8 is recommended). Used to visualise FFEA systems and trajectories, with the plugin we provide.
- NETGEN or TETGEN. Programs which convert surface profile into volumetric meshes to be used by FFEA. Essential if you want to generate meshes from experimental imaging data. We suggest TETGEN!
- Doxygen (>=1.8). Builds the FFEA documentation. Some mathematical formulae will not render correctly if LaTeX is not found.
Optional:
- MDanalysis (>=0.18.0). Used during rod parameterisation. If you don't plan to use KOBRA rods, you can ignore this.
- pyPcazip. Some of the Python FFEA analysis tools interact with the pyPcazip Principal Component Analysis libraries in order to generate standard PCA output (eigensystems, projections, animations etc) equivalent to those obtained from equivalent MD simulations. See here for their wiki.
- GTS (>=0.7.6). The GNU Triangulated Surface Libraries allowing the manipulation and coarsening of surface profiles.
- Meshlab. An open soure system for processing and editing 3D triangular meshes.
Included:
- Boost (>=1.54.0). Used for ease of programming at the initialisation phase. Modules "system", "filesystem" and "program-options" are required. Boost 1.63 is included with FFEA, but this can be adjusted.
- Eigen (>=3.2.10). FFEA uses Eigen to calculate and solve linear approximations to the model i.e. Elastic / Dynamic Network Models. Eigen 3.3.7 will be downloaded by CMake, but this can be adjusted.
- RngStreams. Allows FFEA to safely generate random numbers when running on a number of threads, as well as safe restarts, recovering the state of the random number generators in the last saved time step.
- Tet_a_tet. Used to detect element overlapping in the steric repulsion module.
- mtTkinter (0.4). Used in the PyMOL plugin, allowing safe threading.
Contribute
Do you have a research question that FFEA could help to answer?
- Try FFEA and let us know how you're using the software.
- Send bug reports, questions and feature requests to our issue tracker.
- Fork us and help develop FFEA!
If you have questions and comments, please contact us! For biophysics, contact Sarah Harris ( S.A.Harris@leeds.ac.uk). For finite elements, contact Oliver Harlen (O.G.Harlen@leeds.ac.uk). For KOBRA and stochastic enquiries, contact Daniel Read (D.J.Read@leeds.ac.uk). For software engineering, contact Joanna Leng (J.Leng@leeds.ac.uk).